Help Center
Help Center
News & Updates
1. Newly predicted super-enhancers and enhancers of 11 species based on a more optimized rules.
MACS was used to identify H3K27ac/p300/BRD4 peaks with parameters "-p 1e-5 --keep-dup=auto -w -S --space=50". H3K27ac/p300/BRD4 peaks were used to define the enhancer boundary, followed by further filtering based on the criteria: (1) excluding peaks that overlapped with ENCODE blacklisted genomic regions; and (2) excluding peaks that were located within ±2 kb region of any RefSeq annotated gene promoter. The remaining were defined as enhancers. Then, SEs were identified by using the ROSE (Rank Ordering of Super-Enhancers) algorithm based on the H3K27ac/p300/BRD4 ChIP-seq signal with the default parameters.
2. Data Growth Rate / Summary.
SEA version 3.0 expands the types of Cell/Tissue/Disease from 134 to 260, extends the SEs ranking factors from 1 to 3, increases SEs from 83, 996 to 159526.
3. Functional compositional organization for each SEs.
Researchers have begun to focus on the compositional organization of SEs and its regulated function. Here, peak calling based on Hi-C data was used to identify the functional compositional organization of SEs in different samples of Cell/Tissue/Disease. For convenience, we added "4DGenome" and "SE Constituent" track group to show the chromatin interactions in the target area.
4. Update of the analysis tool "cell-type/tissue/disease specificity of super-enhancer".
We calculated Shannon Entropy for each SE across targeted cell types or tissues, and exhibited the entropy values in the result page.
5. Functional annotation.
Enrichr (http://amp.pharm.mssm.edu/Enrichr/) has been integrated into SEA version 3.0 to process the functional significance analysis of these super enhancers.
6. Conditional query update.
We added "Recognition Factors" and "Searching for Es or SEs" options in the conditional query. The display of query result was adjusted.
MACS was used to identify H3K27ac/p300/BRD4 peaks with parameters "-p 1e-5 --keep-dup=auto -w -S --space=50". H3K27ac/p300/BRD4 peaks were used to define the enhancer boundary, followed by further filtering based on the criteria: (1) excluding peaks that overlapped with ENCODE blacklisted genomic regions; and (2) excluding peaks that were located within ±2 kb region of any RefSeq annotated gene promoter. The remaining were defined as enhancers. Then, SEs were identified by using the ROSE (Rank Ordering of Super-Enhancers) algorithm based on the H3K27ac/p300/BRD4 ChIP-seq signal with the default parameters.
2. Data Growth Rate / Summary.
SEA version 3.0 expands the types of Cell/Tissue/Disease from 134 to 260, extends the SEs ranking factors from 1 to 3, increases SEs from 83, 996 to 159526.
3. Functional compositional organization for each SEs.
Researchers have begun to focus on the compositional organization of SEs and its regulated function. Here, peak calling based on Hi-C data was used to identify the functional compositional organization of SEs in different samples of Cell/Tissue/Disease. For convenience, we added "4DGenome" and "SE Constituent" track group to show the chromatin interactions in the target area.
4. Update of the analysis tool "cell-type/tissue/disease specificity of super-enhancer".
We calculated Shannon Entropy for each SE across targeted cell types or tissues, and exhibited the entropy values in the result page.
5. Functional annotation.
Enrichr (http://amp.pharm.mssm.edu/Enrichr/) has been integrated into SEA version 3.0 to process the functional significance analysis of these super enhancers.
6. Conditional query update.
We added "Recognition Factors" and "Searching for Es or SEs" options in the conditional query. The display of query result was adjusted.
Introduction
1. What is Super-Enhancer Archive?
Super-Enhancer Archive is a web based comprehensive resource focuses on the collection, storage and online analysis of super-enhancers. Our mission is to provide a curated set of information datasets for super-enhancers and tools in multiple genomes, to support and promote research in this area. Especially, we provide a genome-scale landscape to show super-enhancer information in a scalable and flexible manner.
2. Super-enhancers and their importance in gene expression
Super-enhancers are genome regions that are large clusters of transcriptional enhancers and drive expression of genes that define cell identity. The term “super-enhancer” was motioned for the first time by Chen et al. in 2004 (Chen, Yao et al. 2004). They are identified in large-scale by Richard A. Young and his colleagues in 2013(Chapuy, McKeown et al. 2013, Hnisz, Abraham et al. 2013). It has been reported that super-enhancers differ from typical enhancers in size, transcription factor density and content, and the ability to activate transcription (Chapuy, McKeown et al. 2013).
Super-Enhancer Archive is a web based comprehensive resource focuses on the collection, storage and online analysis of super-enhancers. Our mission is to provide a curated set of information datasets for super-enhancers and tools in multiple genomes, to support and promote research in this area. Especially, we provide a genome-scale landscape to show super-enhancer information in a scalable and flexible manner.
2. Super-enhancers and their importance in gene expression
Super-enhancers are genome regions that are large clusters of transcriptional enhancers and drive expression of genes that define cell identity. The term “super-enhancer” was motioned for the first time by Chen et al. in 2004 (Chen, Yao et al. 2004). They are identified in large-scale by Richard A. Young and his colleagues in 2013(Chapuy, McKeown et al. 2013, Hnisz, Abraham et al. 2013). It has been reported that super-enhancers differ from typical enhancers in size, transcription factor density and content, and the ability to activate transcription (Chapuy, McKeown et al. 2013).
Dataset in current release of Super-Enhancer Archive
The current release of SEA incorporates 159,526 super-enhancers that were experimentally (8) and computationally (159,516) identified in 260 cell types/tissues/diseases from four species including Human (104,428 of 143 cell types/tissues/diseases), Mouse (23,969 of 39 cell types/tissues), Drosophila melanogaster (497 of 13 tissues) and Caenorhabditis elegans (131 of three tissues). In addition, SEA stores many super-enhancers related genetic and epigenetic information including sequence conservation, nearby genes, CRISPR-Cas9 target sites, H3K27ac and transcriptional factor binding sites etc.
SEA systematically named each super-enhancer for the first time.
Super-enhancers Naming Convention: Super-enhancers in SEA are named using a specific naming convention. Take underscore as the delimiters, which separate the items clearly.
Super-enhancers Name Format: < reference genome >_< cell-type/tissue/disease >_< number of repeat samples >_< chromosome >_< start site >
| Total | Data acc. | Human | Mouse | D. melanogaster | C. elegans | Zebrafish | Chicken | Chimp | Rhesus | Sheep | X. tropicalis | Stickleback | |||
| Source | Num. | Cell/Tissue/Disease | |||||||||||||
| Super-Enhancer | 159, 308 | SEA (computational) | 164,545 | 266 | 109,447 | 23,969 | 497 | 131 | 2,922 | 5,883 | 5,794 | 13,442 | 887 | 284 | 1,289 |
| SEA (experimental) | 52 | 18 | 52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Recognition factors | 246 | H3K27ac | 210 | 210 | 112 | 30 | 12 | 3 | 10 | 13 | 10 | 13 | 1 | 3 | 3 |
| p300 | 34 | 34 | 26 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| BRD4 | 20 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Med1 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| DNA methylation | 37 | GEO | 37 | 37 | 21 | 15 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Expression | 87 | Roadmap | 87 | 87 | 58 | 14 | 2 | 0 | 4 | 6 | 1 | 1 | 1 | 0 | 0 |
| Reference Genome | 11 | UCSC | 11 | - | hg38 | mm10 | dm6 | ce10 | danRer11 | galGal5 | panTro5 | rheMac8 | oviAri3 | xenTro9 | gasAcu1 |
| CpG Islands | 66, 916 | UCSC | 66, 916 | - | 28, 691 | 16, 026 | 22, 199 | - | x | x | x | x | x | x | x |
| SNP | 4 | UCSC | 4 | 4 | 1 | 1 | 1 | 1 | x | x | x | x | x | x | x |
| Transcription Factor Binding Sites | 1, 104, 318 | JASPAR | 1, 104, 318 | - | 803, 489 | 267, 980 | 13, 357 | 19, 492 | x | x | x | x | x | x | x |
| TF ChIP-seq data | 126 | ENCODE | 126 | 126 | 117 | 9 | 0 | 0 | x | x | x | x | x | x | x |
| CRISPR-Cas9 | 1, 211, 142 | CRISPR Genome Engineering Resources | 1, 211, 142 | - | 518, 266 | 137, 529 | 482, 150 | 73, 197 | x | x | x | x | x | x | x |
Super-enhancers Naming Convention: Super-enhancers in SEA are named using a specific naming convention. Take underscore as the delimiters, which separate the items clearly.
Super-enhancers Name Format: < reference genome >_< cell-type/tissue/disease >_< number of repeat samples >_< chromosome >_< start site >
| Label | Description |
|---|---|
| reference genome | The beginning of the name is the version number of a super-enhancer’s reference genome, for example, "mm9" is the reference genome version of super-enhancer mm9_BAT_1_chr9_3002875. |
| cell-type/tissue/disease | The cell-type/tissue/disease is the sample which the super-enhancer was discovered in. It is represented by an abbreviation we defined. For example, "BAT" is the tissue type of super-enhancer mm9_BAT_1_chr9_3002875, and it is the short for Brown adipocytes tissue. |
| number of repeat samples | Because each cell-type/tissue/disease contain one or more samples, an Arabic numeral was used to distinguish them, the number is omitted if there is only one sample involved to this tissue/cell. |
| chromosome | The fourth item is the chromosome number of a super-enhancer. |
| start site | The last is the start site of a super-enhancer in genome location. |
Features of Super-Enhancer Archive
As a specifically designed super-enhancer database, SEA not only could integrate the dispersed data but also provide a convenient way for in-depth data mining. In details,
(i) stores comprehensive super-enhancers of four species including human, mouse and other two model species (Drosophila melanogaster and Caenorhabditis elegans) supporting further analysis of their sequence conservation, and the number of species would be increased when the needed data emerge;
(ii) displays the cell-type/tissue/disease specificity of super-enhancer by comparative analysis by location, which is a key feature of super-enhancer and useful for analysis of their cell-type/tissue/disease specific regulatory function;
(iii) identifies the relationships among genes and super-enhancer by flexible analysis of online regulatory network induced by super-enhancer;
(iv) provides the CRISPR-Cas9 target sites computationally predicted by SSfinder in super-enhancers which is useful for further function study and application via genome edition;
(v) supplements classification of publications related to super-enhancers according research content, which should be useful for biologist following advance of super-enhancer study;
(vi) offers two online analysis tools, one for genomic regions enrichment analysis and another for H3K27ac cell-type-specificity analysis;
(vii) develops a genome browser and supports customized and user-friendly views in genomic scale. The gene-centric super-enhancer information together with genomic and epigenetic information (such as H3K27ac and transcriptional factor binding sites) are available to view in the browser. The browser can link out to other databases such as HHMD, DiseaseMeth, MetaImprint and DevMouse.
(i) stores comprehensive super-enhancers of four species including human, mouse and other two model species (Drosophila melanogaster and Caenorhabditis elegans) supporting further analysis of their sequence conservation, and the number of species would be increased when the needed data emerge;
(ii) displays the cell-type/tissue/disease specificity of super-enhancer by comparative analysis by location, which is a key feature of super-enhancer and useful for analysis of their cell-type/tissue/disease specific regulatory function;
(iii) identifies the relationships among genes and super-enhancer by flexible analysis of online regulatory network induced by super-enhancer;
(iv) provides the CRISPR-Cas9 target sites computationally predicted by SSfinder in super-enhancers which is useful for further function study and application via genome edition;
(v) supplements classification of publications related to super-enhancers according research content, which should be useful for biologist following advance of super-enhancer study;
(vi) offers two online analysis tools, one for genomic regions enrichment analysis and another for H3K27ac cell-type-specificity analysis;
(vii) develops a genome browser and supports customized and user-friendly views in genomic scale. The gene-centric super-enhancer information together with genomic and epigenetic information (such as H3K27ac and transcriptional factor binding sites) are available to view in the browser. The browser can link out to other databases such as HHMD, DiseaseMeth, MetaImprint and DevMouse.
Querying & Browsing
Accurate query entrance is provided for users to search the super-enhancer information for their concerned genome locations, genes or cell-type/tissue/diseases.
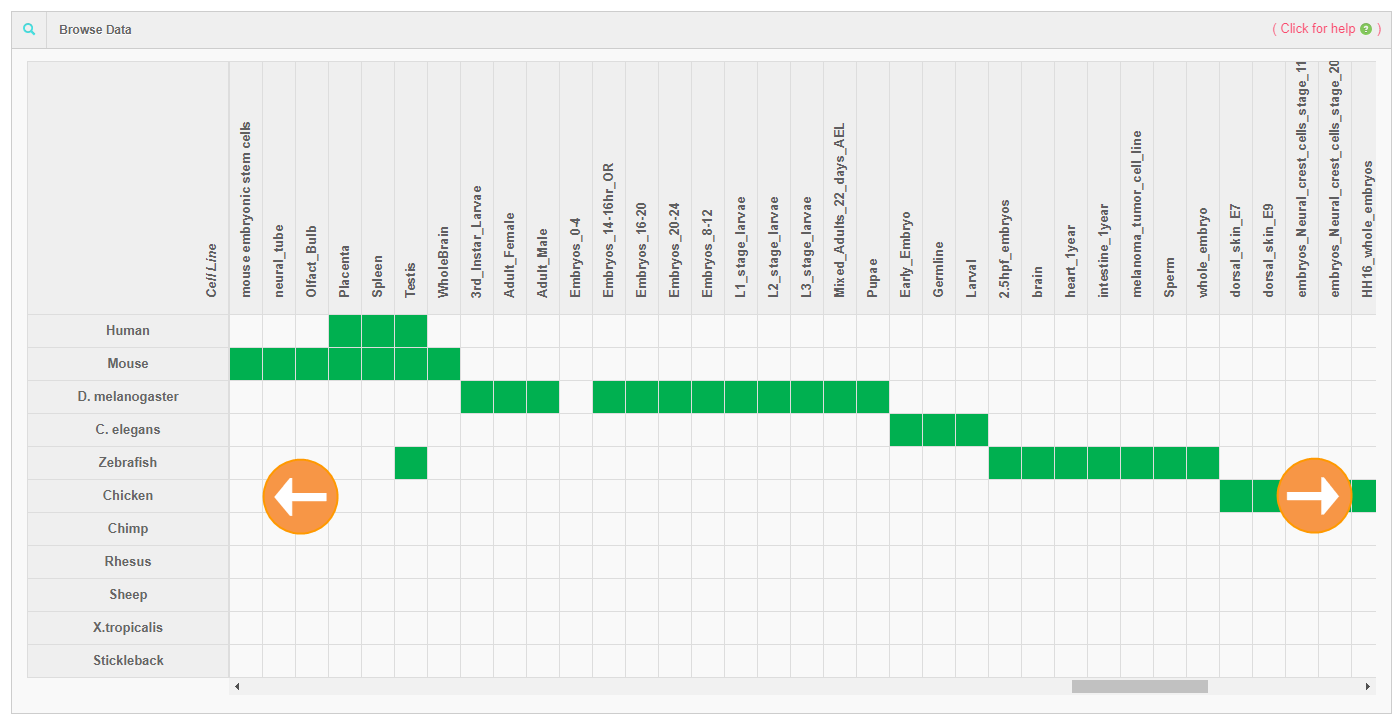
1. Table browser for datasets
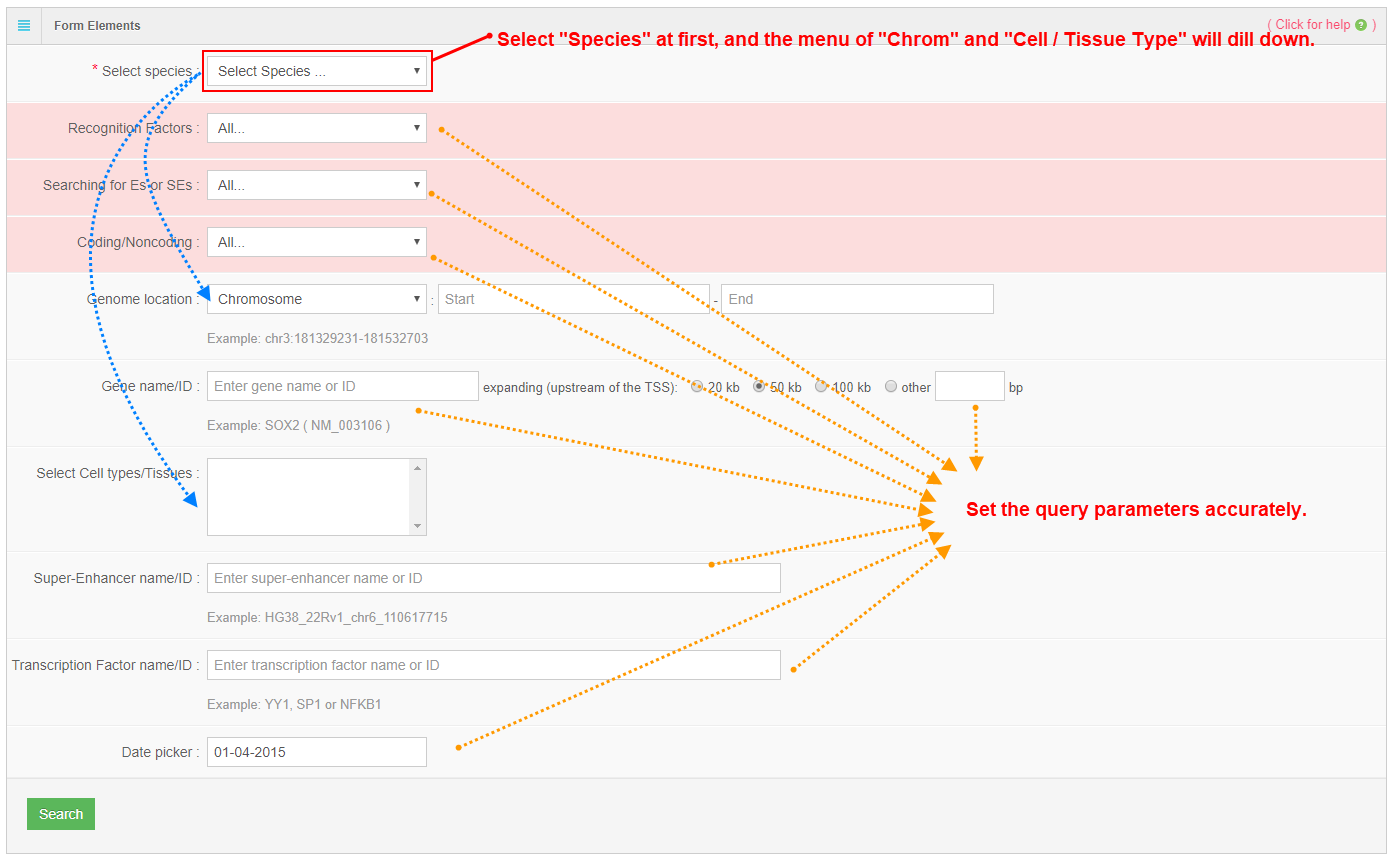
2. Advance search
In this search entrance, accurate search is supported. User can specify the detailed information about super-enhancers such as species, genome location, gene name, cell-type/tissue, super-enhancer name, transcription factor etc. The results would be showed in the tables and SEA-browser.
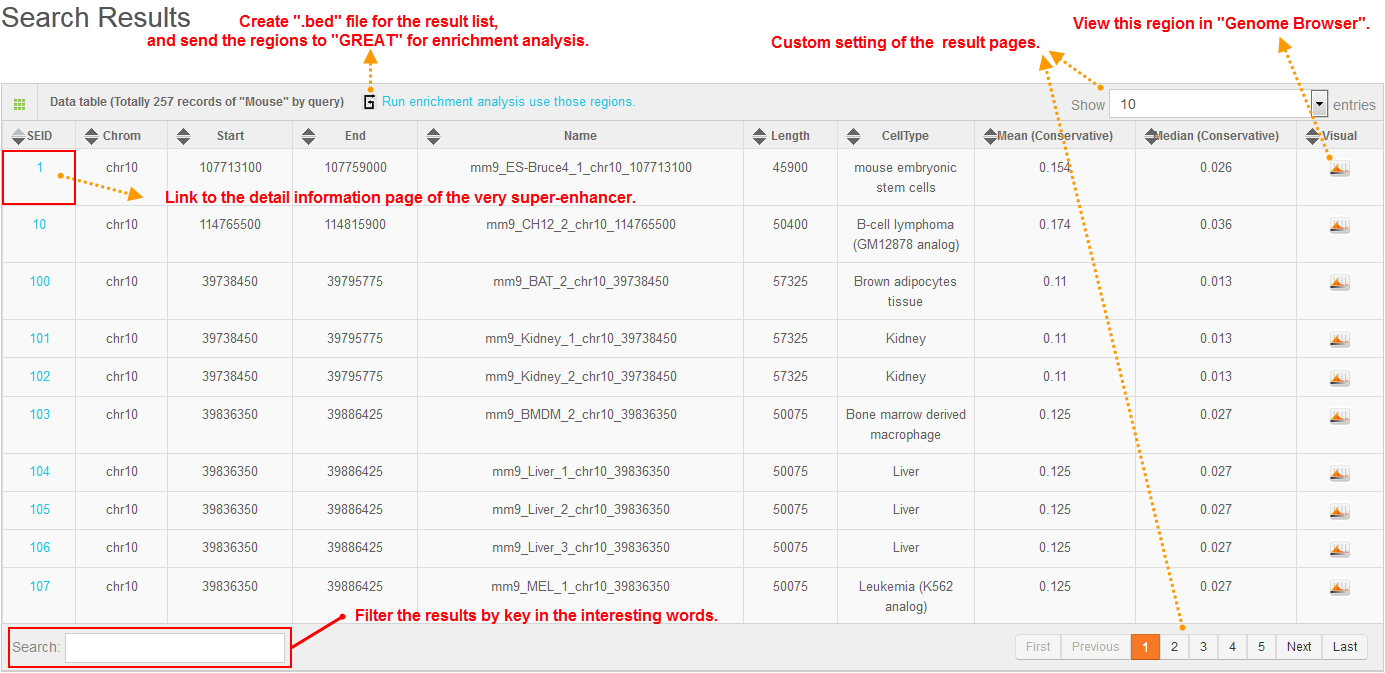
3. Search result table
The search result would be shown in table. User can see the detail information by the links listed in this table, such as the SEID for the detailed information about nearest gene, cell-type specificity and sequence conservation. The links to SEA-browser would bring to SEA-browser for visualization of these information in genomic context.
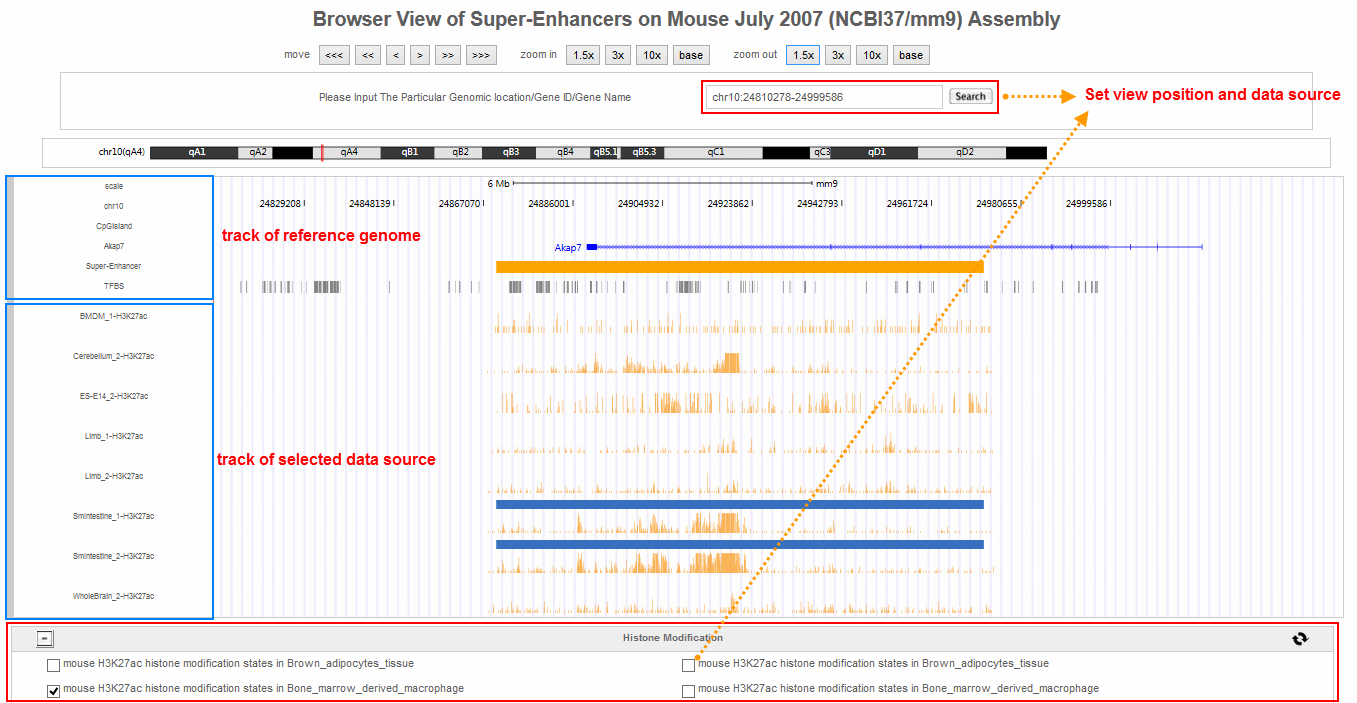
4. SEA-browser
SEA-browser was developed for visualization of super-enhancer information in the genomic context. In SEA-browser, user can see the location of super-enhancer to nearby genes, transcriptional factor binding sites and the cell-type-specific super-enhancer and H3K27ac modification pattern. User can add or remove the data tracks by clicking the selector at the bottom of this pape.
5. A case study for analysis of human gene SOX2
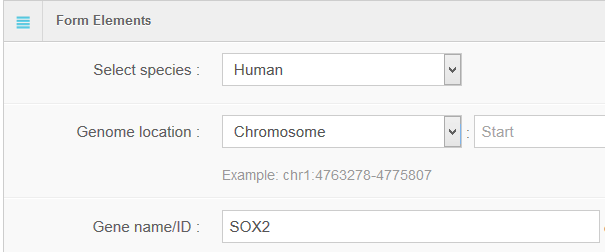
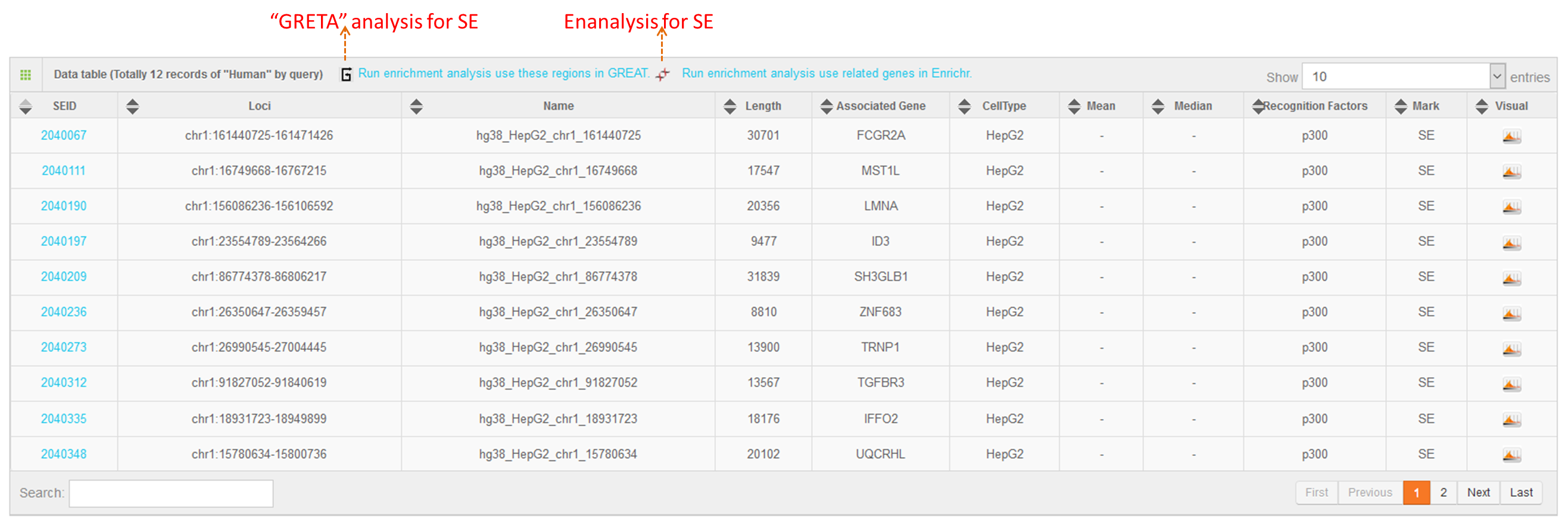
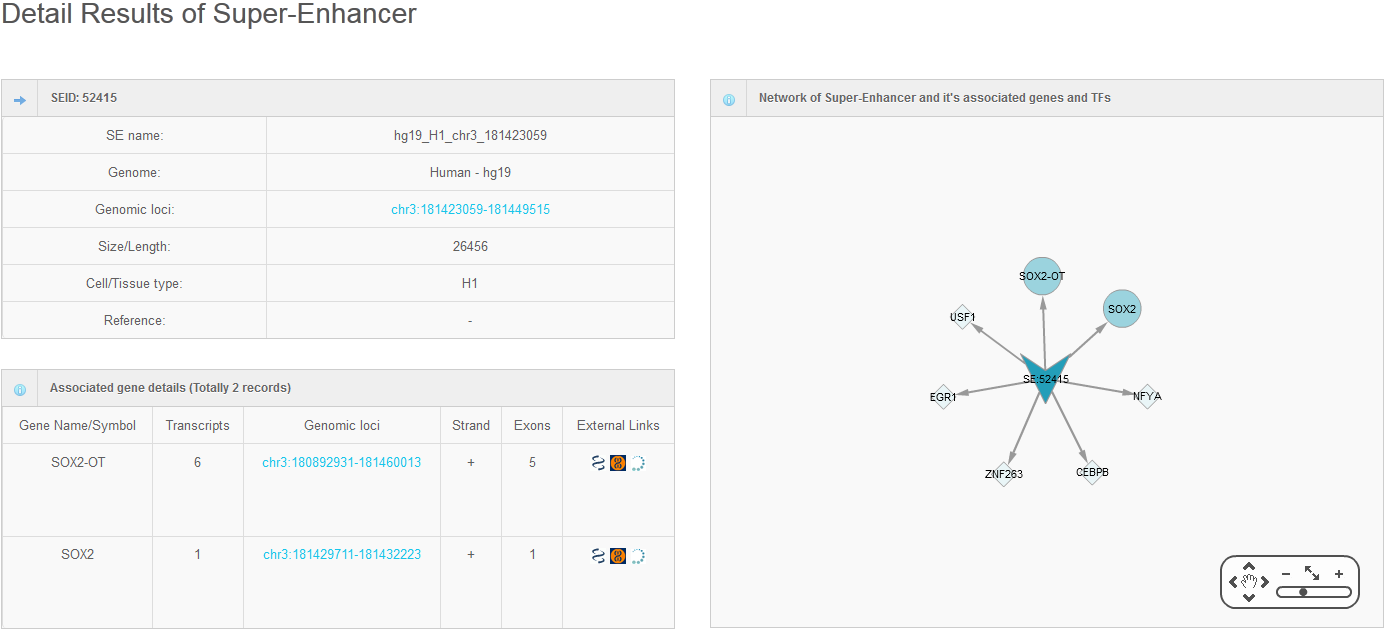
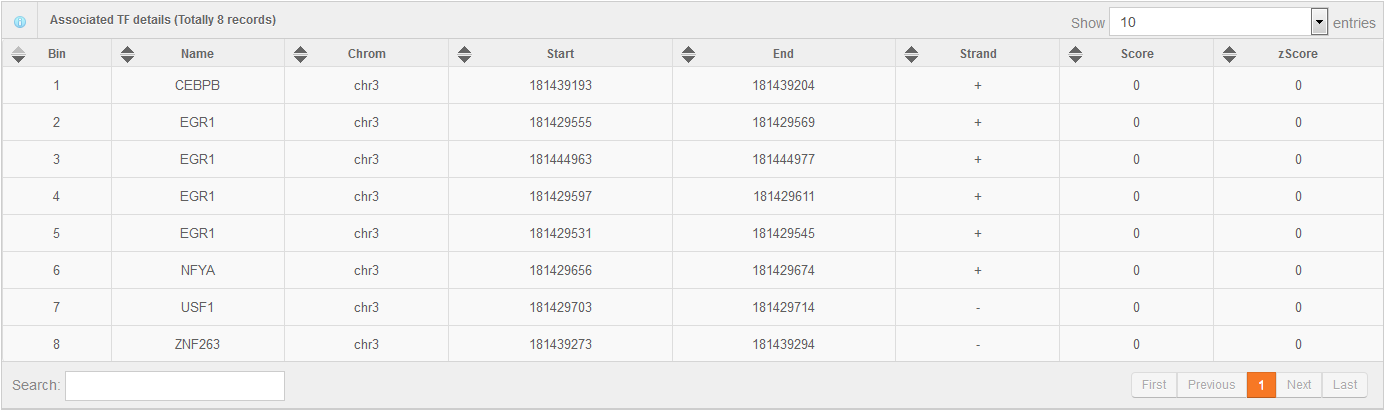
Hello every one, I am Hongbo Liu. I am interested in the super-enhancer related to human gene SOX2 which coding an important transcript factor for stem cell pluripotent. In the Advance Search page, I selected human species and input “SOX2” as gene name. After clicking the Search button on the bottom, eight super-enhancers related with human SOX2 in different cell-type/tissues are list in the search result page. It is well known that SOX2 is a key gene for stem cell. Thus, I selected the super-enhancer (SEID:30459) in human stem cell H1. Clicking the blue colored SEID, the detailed information about this super-enhancer is shown in a detailed information page. As shown in this page, there are two genes (SOX2 and SOX2-OT) nearby this super-enhancer. From the network and table, we acquired that there are 5 transactional factor binding site (including the well-known super-enhancer marks CEBP and MYOD) in this super-enhancer. In order to examine the relationship between this super-enhancer and gene SOX2, I go to the SEA-Browser via the links provided in the search result page or the detailed information page. In the SEA-Browser, the genetic and epigenetic context in the super-enhancer are visualized. It is shown that gene SOX2 is localized in this super-enhancer. This super-enhancer includes three CpG islands which are key regulatory regions for DNA methylation. As an important feature of super-enhancer, this super-enhancer contains a lot of transcriptional factor binding sites which are essential for the transcription regulation of gene SOX2. It should be noted that SEA-Browser provides the super-enhancer and H3K27ac modification state in all cell-type/tissue/diseases which enable us to check the cell-type/tissue/disease specific super-enhancer and H3K27ac modification. It is shown that this super-enhancer is stem cell specific and specifically enriched by H3K27ac, indicating the important roles of this super-enhancer in regulation of stem cell specifically high expression of gene SOX2. In addition, search result also provide me useful links to other gene annotation databases including NCBI Gene, GeneCards and UniProt, by which I learned more information about human gene SOX2. In short, I experienced a very good experience in SEA, and also learned new knowledge about gene SOX2 and its super-enhancer. Well Done! I hope you will like this database!

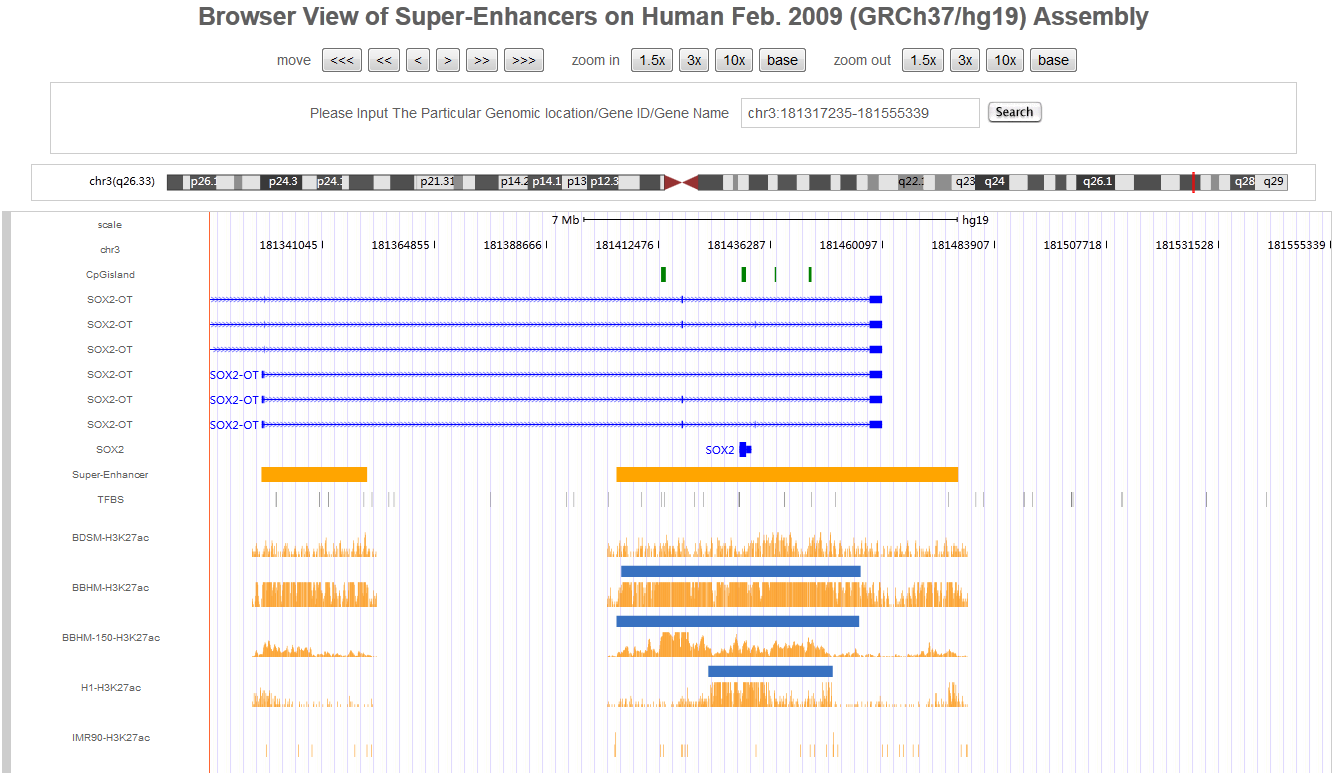
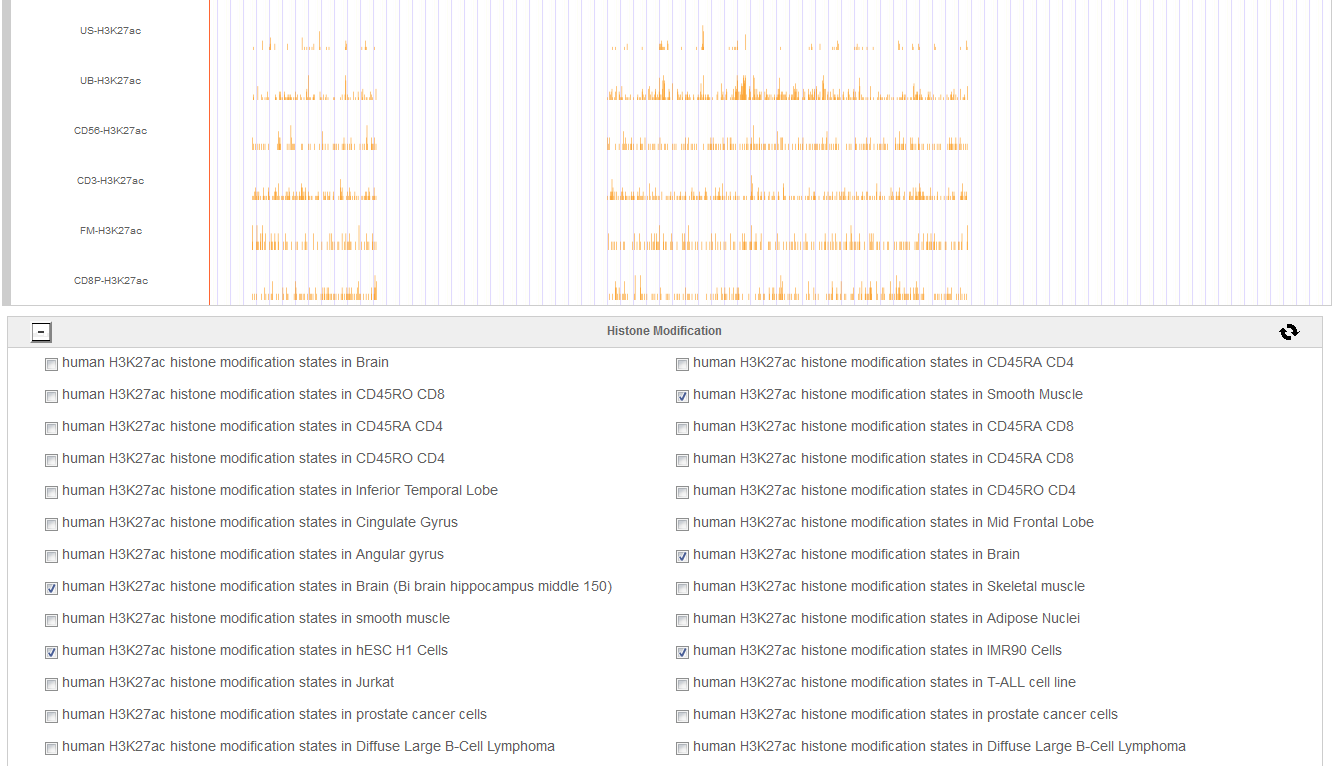
6. Online analysis tools for super-enhancers
Using the advanced search for super-enhancers, users can obtain a “.bed” file (Figure 1B), which can be downloaded for future study or sent to the GREAT server (http://bejerano.stanford.edu/great/public/html/) for genomic enrichment of annotations.
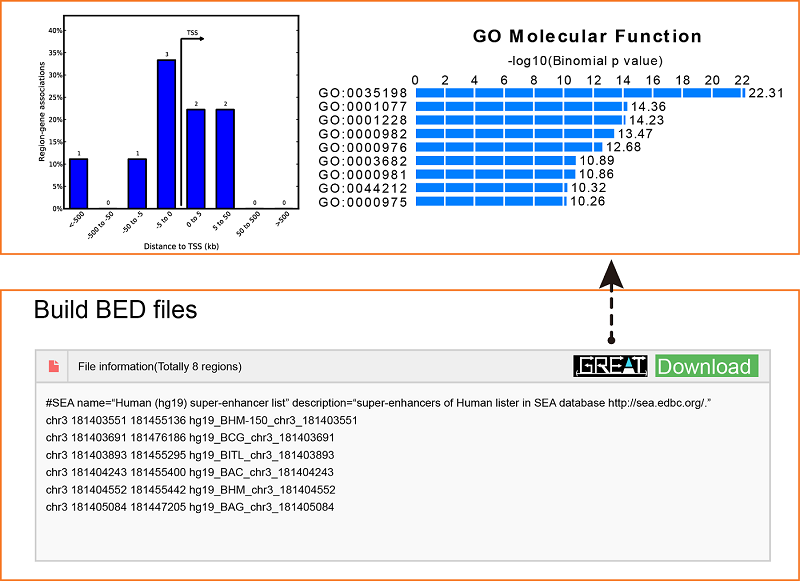
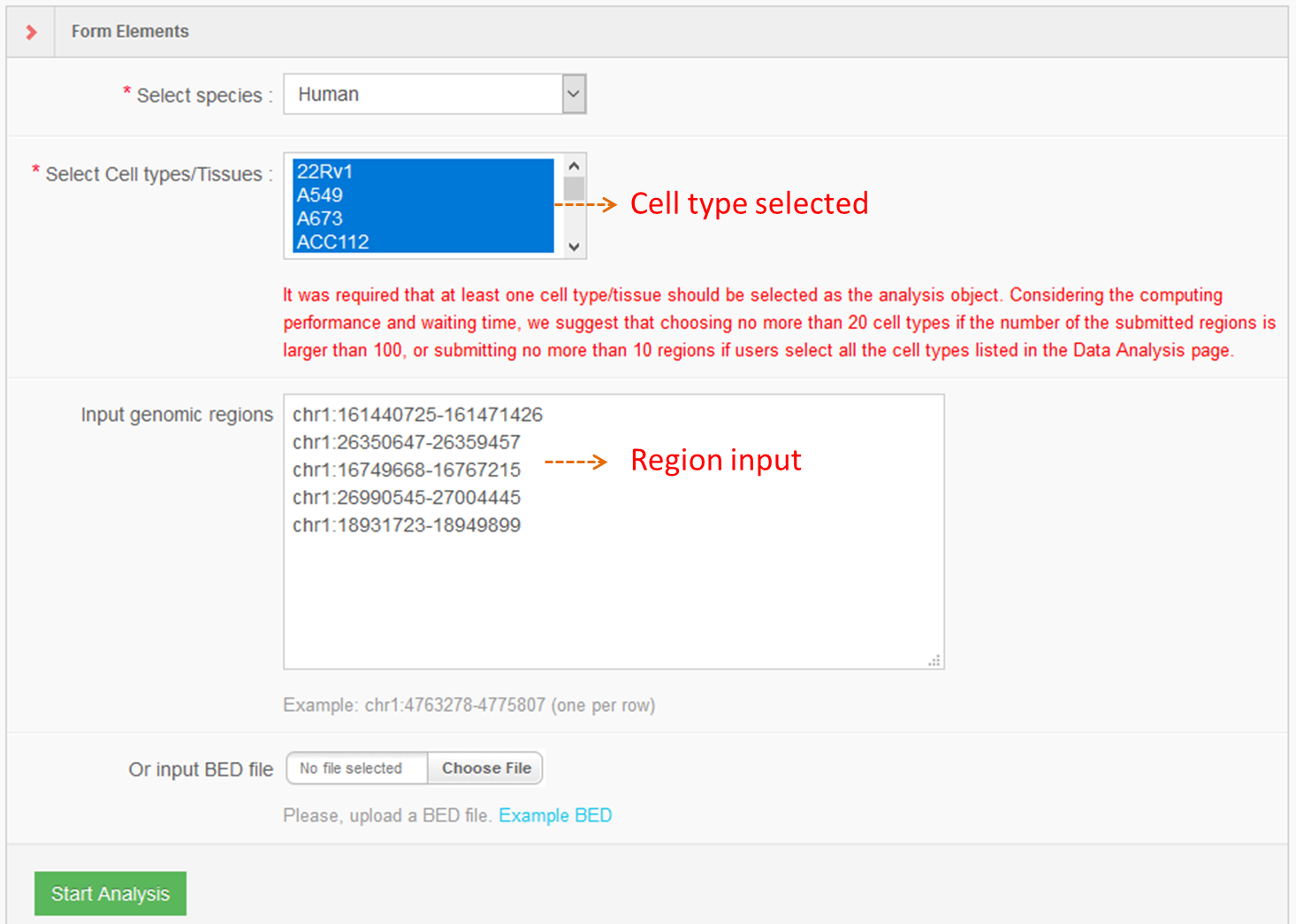
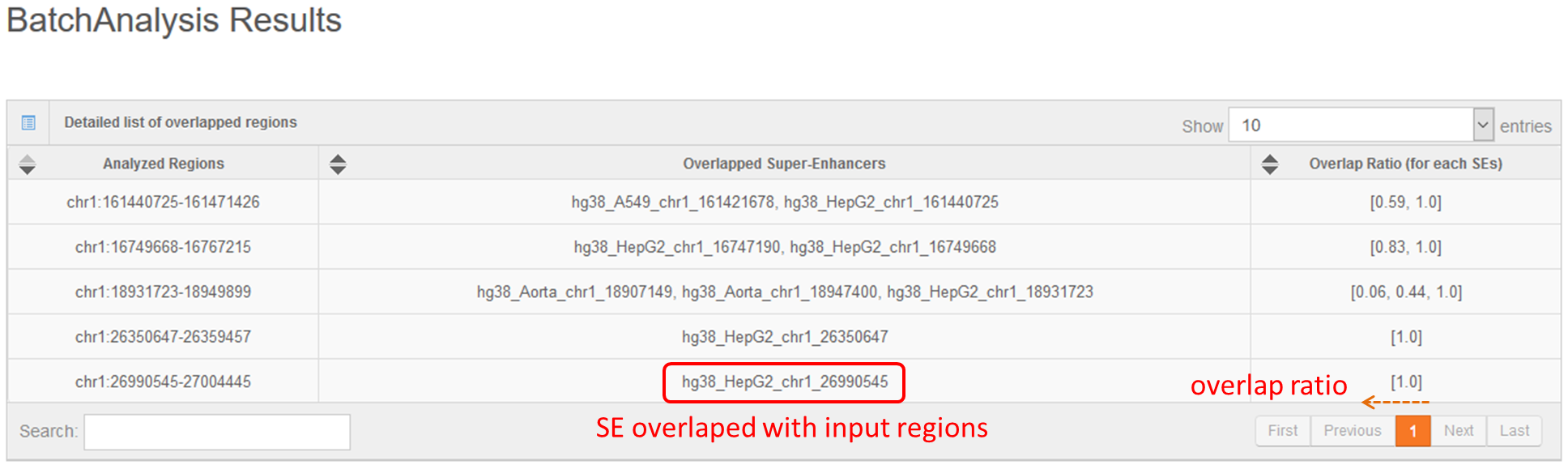
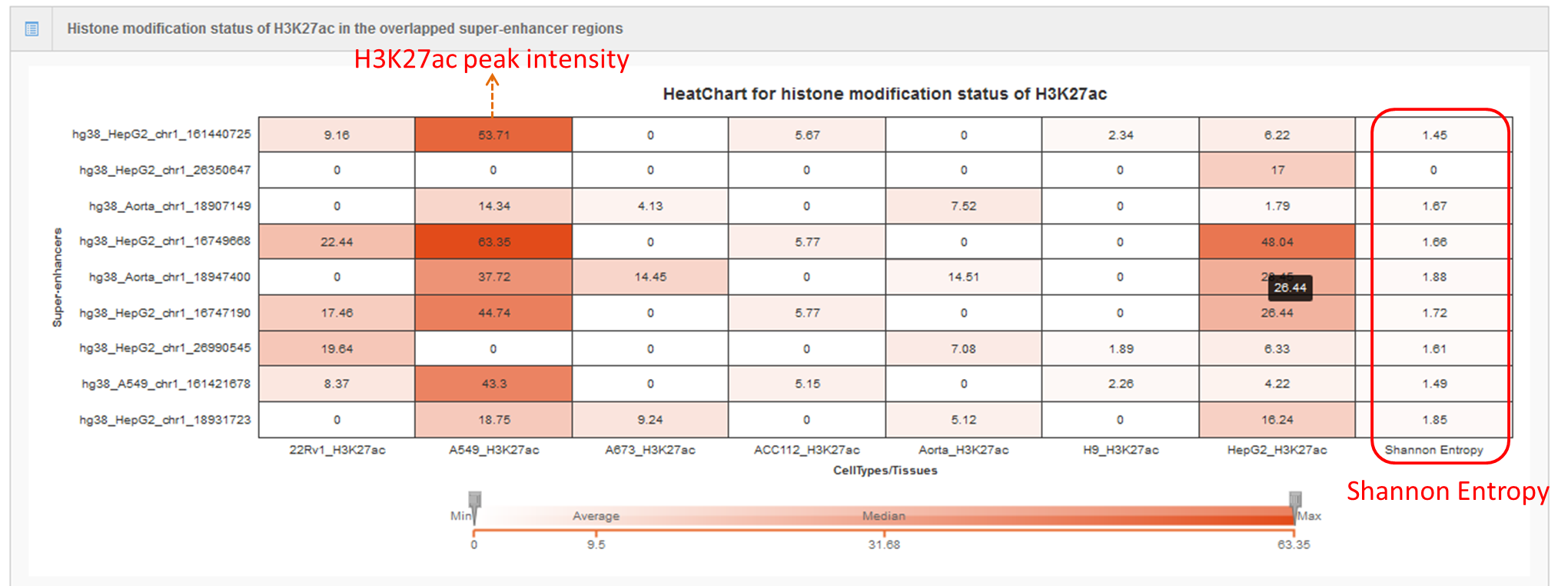
Users can also submit the genomic region(s) to SEA, which will calculate and list the overlap of the regions of interest with the current super-enhancer regions in the SEA database for downstream analysis. Simultaneously, the back-end will calculate the average H3K27ac status for each overlapping super-enhancer, and present the super-enhancer histone modification level in different cell types/tissues/diseases in a heat map.Lastly, Shannon Entropy is calculated for every SE across selected cell types. The more the value close to 0, the more specificity of the SE across selected cell types.
7. Format specification for Custom Tracks
< position > representing the initial regions for visualization.
< track > for define the custom tracks' name and detailed description.
< color > for define the custom tracks' display color.
< type > for define the custom tracks' display style, "hist" will show the custom data as peak value diagram and "region" will show the custom data as square chart.
Example 1:
OR
Example 2:
1. Table browser for datasets

2. Advance search
In this search entrance, accurate search is supported. User can specify the detailed information about super-enhancers such as species, genome location, gene name, cell-type/tissue, super-enhancer name, transcription factor etc. The results would be showed in the tables and SEA-browser.

3. Search result table
The search result would be shown in table. User can see the detail information by the links listed in this table, such as the SEID for the detailed information about nearest gene, cell-type specificity and sequence conservation. The links to SEA-browser would bring to SEA-browser for visualization of these information in genomic context.

4. SEA-browser
SEA-browser was developed for visualization of super-enhancer information in the genomic context. In SEA-browser, user can see the location of super-enhancer to nearby genes, transcriptional factor binding sites and the cell-type-specific super-enhancer and H3K27ac modification pattern. User can add or remove the data tracks by clicking the selector at the bottom of this pape.

5. A case study for analysis of human gene SOX2
Hello every one, I am Hongbo Liu. I am interested in the super-enhancer related to human gene SOX2 which coding an important transcript factor for stem cell pluripotent. In the Advance Search page, I selected human species and input “SOX2” as gene name. After clicking the Search button on the bottom, eight super-enhancers related with human SOX2 in different cell-type/tissues are list in the search result page. It is well known that SOX2 is a key gene for stem cell. Thus, I selected the super-enhancer (SEID:30459) in human stem cell H1. Clicking the blue colored SEID, the detailed information about this super-enhancer is shown in a detailed information page. As shown in this page, there are two genes (SOX2 and SOX2-OT) nearby this super-enhancer. From the network and table, we acquired that there are 5 transactional factor binding site (including the well-known super-enhancer marks CEBP and MYOD) in this super-enhancer. In order to examine the relationship between this super-enhancer and gene SOX2, I go to the SEA-Browser via the links provided in the search result page or the detailed information page. In the SEA-Browser, the genetic and epigenetic context in the super-enhancer are visualized. It is shown that gene SOX2 is localized in this super-enhancer. This super-enhancer includes three CpG islands which are key regulatory regions for DNA methylation. As an important feature of super-enhancer, this super-enhancer contains a lot of transcriptional factor binding sites which are essential for the transcription regulation of gene SOX2. It should be noted that SEA-Browser provides the super-enhancer and H3K27ac modification state in all cell-type/tissue/diseases which enable us to check the cell-type/tissue/disease specific super-enhancer and H3K27ac modification. It is shown that this super-enhancer is stem cell specific and specifically enriched by H3K27ac, indicating the important roles of this super-enhancer in regulation of stem cell specifically high expression of gene SOX2. In addition, search result also provide me useful links to other gene annotation databases including NCBI Gene, GeneCards and UniProt, by which I learned more information about human gene SOX2. In short, I experienced a very good experience in SEA, and also learned new knowledge about gene SOX2 and its super-enhancer. Well Done! I hope you will like this database!







6. Online analysis tools for super-enhancers
Using the advanced search for super-enhancers, users can obtain a “.bed” file (Figure 1B), which can be downloaded for future study or sent to the GREAT server (http://bejerano.stanford.edu/great/public/html/) for genomic enrichment of annotations.

Users can also submit the genomic region(s) to SEA, which will calculate and list the overlap of the regions of interest with the current super-enhancer regions in the SEA database for downstream analysis. Simultaneously, the back-end will calculate the average H3K27ac status for each overlapping super-enhancer, and present the super-enhancer histone modification level in different cell types/tissues/diseases in a heat map.Lastly, Shannon Entropy is calculated for every SE across selected cell types. The more the value close to 0, the more specificity of the SE across selected cell types.



7. Format specification for Custom Tracks
< position > representing the initial regions for visualization.
< track > for define the custom tracks' name and detailed description.
< color > for define the custom tracks' display color.
< type > for define the custom tracks' display style, "hist" will show the custom data as peak value diagram and "region" will show the custom data as square chart.
Example 1:
position chr3:181403551-181455442
track name="test peaks" description="super-enhancers of Huamn listed in SEA database http://sea.edbc.org/."
color #6600ff
type region
chr3 181403551 181455136 regin name 1
chr3 181403691 181476186 region name 2
OR
Example 2:
position chr3:181403551-181455442
track name="test peaks" description="super-enhancers of Huamn listed in SEA database http://sea.edbc.org/."
color #6600ff
type hist
chr3 181403551 181455136 0.1
chr3 181403691 181476186 0.3
Materials & Methods
Identification of Super-enhancers based on H3K27ac / p300 / BRD4
Currently, due to the large size of super-enhancers, it is difficult and time-consuming to find novel super-enhancers in a mount of cell-types/tissues/diseases, although there are a few super-enhancers identified based on experiment. Thus, Candidate super-enhancers are mainly identified by computational method, such as the ROSE program which was developed for identification of super-enhancers based H3K27ac ChIP-Seq dataset by Richard A. Young and his colleagues. Recently, there are a lot of epigenome projects (such as ENCODE, modENCODE, and Human Epigenome Roadmap) aimed at profiling H3K27ac, which is regarded as the key epigenetic indicator for super-enhancer in various cell-types, normal tissues and diseases. Wide profiling of H3K27ac in many cell-type/tissue/diseases raised the possibility to integrate these datasets to systematically identify specific super-enhancers in cell-type/tissue/disease. In this study, we took full advantage of the cross-dataset analysis of cell-type/tissue/disease specific super-enhancers to discover potentially novel genes/regions in cell-type/tissue/diseases for biologists. In total, we integrated the H3K27ac profiles together and used ROSE program to predict a comprehensive map of super-enhancers in four model species (Human, Mouse, Drosophila melanogaster, and Caenorhabditis elegans) whose H3K27ac dataset are available from public data resource.
Reference classification and collection of experimentally confirmed super-enhancers
To mine the super-enhancers from the literature, we searched the PubMed database using the keywords “super enhancer” and obtained 138 publications. We then scanned each publication and removed 94 that were not gene-related studies. From the remaining 44 publications, we manually classified each study into seven clusers including Development, Cancer, Disease, Transcriptional regulation, Genome editing with CRISPR-Cas9, Human, and Mouse according to its research content. Further, we excavated experimentally confirmed super-enhancers according to the description of the experiment in each study. Especially, we focused on the collection of response after genome editing in super-enhancer with CRISPR-Cas9. For example, using a simple and highly efficient double-CRISPR genome editing strategy, Bing Ren and his colleague deleted a entire 13-kb super-enhancer located 100kb downstream of Sox2 in mouse ESCs and characterized transcriptional defects in the resulting monoallelic and biallelic deletion clones with RNA-seq (Li, Rivera et al. 2014).
Identification of nearby genes for each super-enhancer
It has been reported super-enhancer participates in regulating the cell-type-specific expression of its nearest gene. In the search engine, the nearest gene for each super-enhancer would be identified automatically based on location. More over, the super-enhancers nearby a given gene also can be identified automatically with the expanding length (default 50kb) can be set up by users.
Currently, due to the large size of super-enhancers, it is difficult and time-consuming to find novel super-enhancers in a mount of cell-types/tissues/diseases, although there are a few super-enhancers identified based on experiment. Thus, Candidate super-enhancers are mainly identified by computational method, such as the ROSE program which was developed for identification of super-enhancers based H3K27ac ChIP-Seq dataset by Richard A. Young and his colleagues. Recently, there are a lot of epigenome projects (such as ENCODE, modENCODE, and Human Epigenome Roadmap) aimed at profiling H3K27ac, which is regarded as the key epigenetic indicator for super-enhancer in various cell-types, normal tissues and diseases. Wide profiling of H3K27ac in many cell-type/tissue/diseases raised the possibility to integrate these datasets to systematically identify specific super-enhancers in cell-type/tissue/disease. In this study, we took full advantage of the cross-dataset analysis of cell-type/tissue/disease specific super-enhancers to discover potentially novel genes/regions in cell-type/tissue/diseases for biologists. In total, we integrated the H3K27ac profiles together and used ROSE program to predict a comprehensive map of super-enhancers in four model species (Human, Mouse, Drosophila melanogaster, and Caenorhabditis elegans) whose H3K27ac dataset are available from public data resource.
Reference classification and collection of experimentally confirmed super-enhancers
To mine the super-enhancers from the literature, we searched the PubMed database using the keywords “super enhancer” and obtained 138 publications. We then scanned each publication and removed 94 that were not gene-related studies. From the remaining 44 publications, we manually classified each study into seven clusers including Development, Cancer, Disease, Transcriptional regulation, Genome editing with CRISPR-Cas9, Human, and Mouse according to its research content. Further, we excavated experimentally confirmed super-enhancers according to the description of the experiment in each study. Especially, we focused on the collection of response after genome editing in super-enhancer with CRISPR-Cas9. For example, using a simple and highly efficient double-CRISPR genome editing strategy, Bing Ren and his colleague deleted a entire 13-kb super-enhancer located 100kb downstream of Sox2 in mouse ESCs and characterized transcriptional defects in the resulting monoallelic and biallelic deletion clones with RNA-seq (Li, Rivera et al. 2014).
Identification of nearby genes for each super-enhancer
It has been reported super-enhancer participates in regulating the cell-type-specific expression of its nearest gene. In the search engine, the nearest gene for each super-enhancer would be identified automatically based on location. More over, the super-enhancers nearby a given gene also can be identified automatically with the expanding length (default 50kb) can be set up by users.
FAQs
1. Why the gene information is unavailable sometimes?
A: The genomic region you choose is too wide, it would be too slow to show all of the RefSeq genes.
2. There is a long time to wait for querying, why?
A: If waiting extremely a long time, user should take the network delay into consideration, though there is the possibility that the server encounters many concurrent requests. It is possible that the super-enhancers involved for a specific search conditions are too many. If user encounters potential common problems, click to go to help page.
3. What methods are used to identify the Super-Enhancers?
A: The genomic region you choose is too wide, it would be too slow to show all of the RefSeq genes.
2. There is a long time to wait for querying, why?
A: If waiting extremely a long time, user should take the network delay into consideration, though there is the possibility that the server encounters many concurrent requests. It is possible that the super-enhancers involved for a specific search conditions are too many. If user encounters potential common problems, click to go to help page.
3. What methods are used to identify the Super-Enhancers?
| Researches | Methods | Factors | Parameters |
| Loven et al. | ROSE | Med1 | stitching distance 12,500, promoter exclusion 2,500 |
| Hnisz et al. | ROSE | H3K27ac | stitching distance 12,500, promoter exclusion2,000 |
| Kwiatkowski et al. | ROSE | H3K27ac | stitching distance 12,500 |
| SEA | ROSE | H3K27ac / p300 / BRD4 | stitching distance 12,500, promoter exclusion2,000 |