Site Analytics
Data Growth Rate / Summary
|
45%
|
62%
|
|
67%
|
6.5%
|
- Flow Chart
- Introduction
- Super-Enhancer
- Highlight
SEA version 3.0 (i) stores the newly predicted super-enhancers and enhancers of 11 species based on a more optimized rules, expands the types of Cell/Tissue/Disease from 134 to 246, extends the SEs ranking factors from 1 to 3 and the number of species would be increased when the needed data emerge; (ii) includes experimentally identified and confirmed super-enhancers; (iii) lists the functional compositional organization for each SEs through Hi-C data based peak calling; (iv) provides the cell-type/tissue/disease specificity of super-enhancer with an quantitative entropy value; (v) includes online regulatory network supporting flexible analysis of the relationships among coding / non-coding genes and super-enhancer; (vi) supplements classification of publications related to super-enhancers which should be useful for biologist following advance of super-enhancer research; (vii) updated the visualization tool “Browser View of Super-Enhancers”, which is contained of abundant annotation tracks with supporting customized views in genomic scale; (viii) integrates Enrichr into SEA version 3.0 to process the functional significance analysis of these super enhancers.
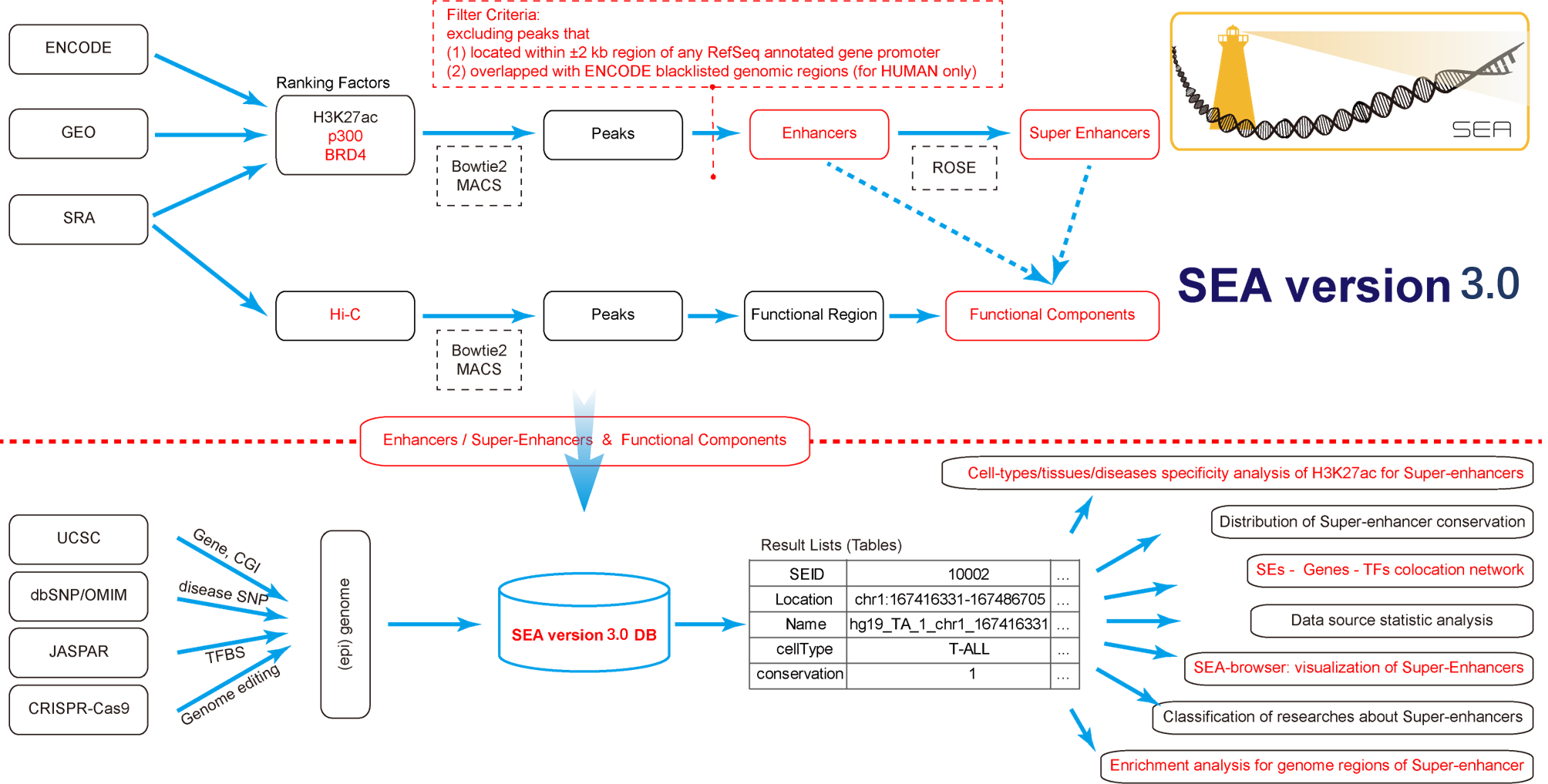
Super-Enhancer Archive is a web based comprehensive resource focuses on the collection, storage and online analysis of super-enhancers. Our mission is to provide a curated set of information datasets for super-enhancers and tools in mutiple genomes, to support and promote research in this area. Especially, we provide a genome-scale landscape to show super-enhancer information in a scalable and flexible manner.
Currently, Super-Enhancer Archive consists of super-enhancers in 11 organisms, including 104,428 human super-enhancers, 23, 969 mouse super-enhancers, 497 D. melanogaster super-enhancers, 131 C. elegans super-enhancers, 2, 922 Zebrafish super-enhancers, 5, 883 Chicken super-enhancers, 5, 794 Chimp super-enhancers, 13, 442 Rhesus super-enhancers, 887 Sheep super-enhancers, 284 X. tropicalis super-enhancers and 1, 289 Stickleback super-enhancers respectively. Super-Enhancer Archive supports analysis and chart view of these datasets, download for further analysis and can be even easily visualized in the genome browser (SEA-Browser).
Further information of citation, updates and statistics are available in this page. This site is updating regularly.
Super-enhancers are genome regions that are large clusters of transcriptional enhancers and drive expression of genes that define cell identity. The term “super-enhancer” was motioned for the first time by Chen et al. in 2004 (Chen, Yao et al. 2004). They are identified in large-scale by Richard A. Young and his colleagues in 2013 (Chapuy, McKeown et al. 2013, Hnisz, Abraham et al. 2013). It has been reported that super-enhancers differ from typical enhancers in size, transcription factor density and content, and the ability to activate transcription (Chapuy, McKeown et al. 2013). And Super-enhancers are regarded as the regulators of cell identity, ESC state maintenance, and tumor pathogenesis.
For more information about super-enhancers, please go to the Documentation page in which the studies about super-enhancers are subtotaled.
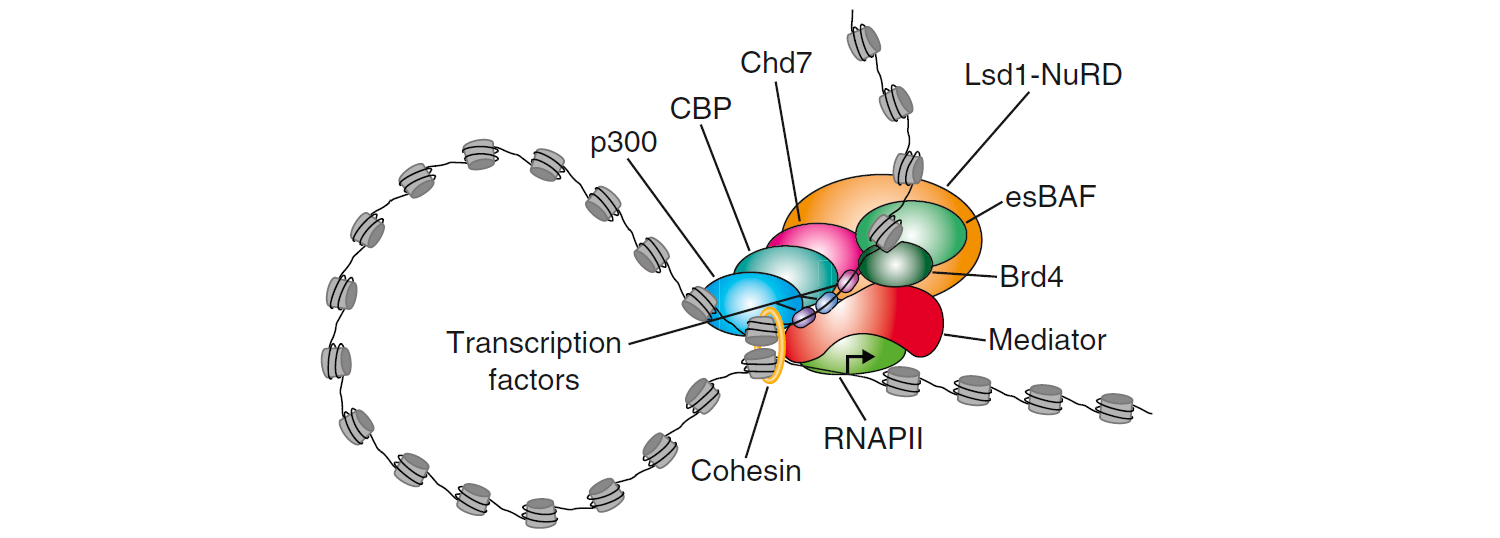
(Hnisz, D., Abraham, B.J., Lee, T.I., Lau, A., Saint-Andre, V., Sigova, A.A., Hoke, H.A. and Young, R.A. (2013) Super-enhancers in the control of cell identity and disease. Cell, 155, 934-947.)
New - Super-enhancers have been newly predicted in 11 species based on a more optimized rules and ChIP-seq data of 3 different ranking factors.
Uptodate - It contains the functional compositional organization for each SEs through Hi-C data based peak calling.
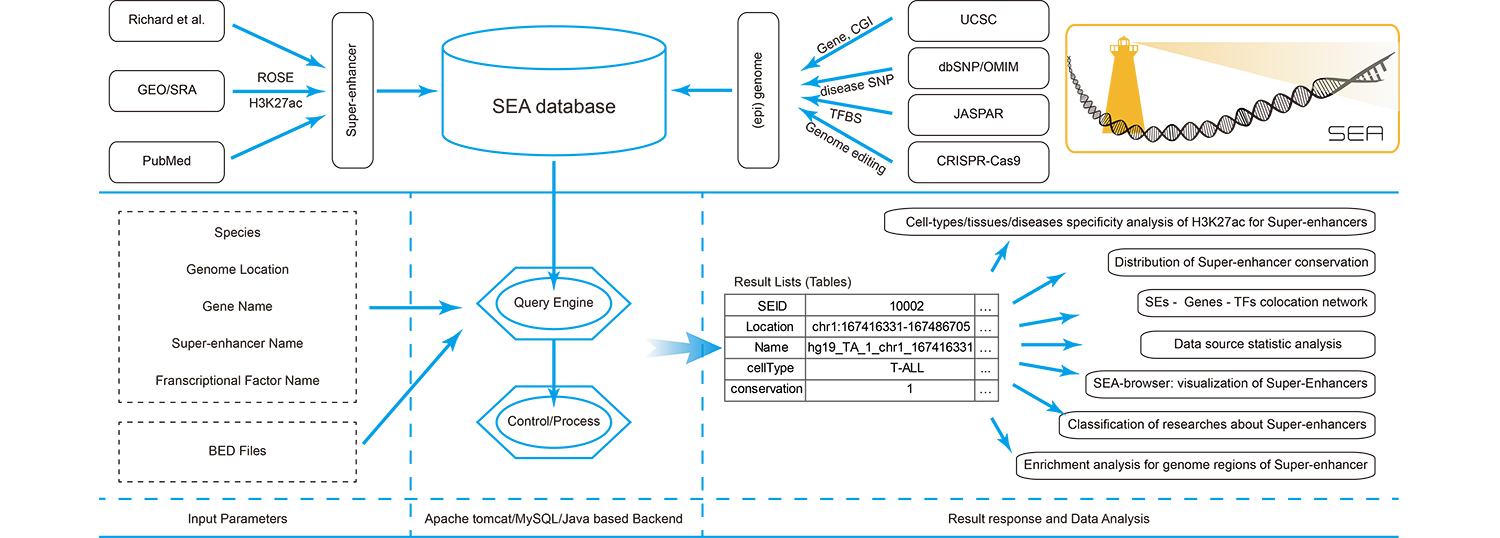
Convenient - It is convenient for biologists to query super-enhancer and its' related information of interested topics.
Functional - Function annotation could be done by integrated online analysis tools (SEA-Browser, TFBS enrichment, H3K27ac enrichment, Enrichr and GREAT).

Super-Enhancers have crucial functions in genome, just like the lighthouse illuminating the road ahead, they drive cell-type-specific extremely high expression of genes that define cell identity in development and disease.