
Best experienced using the browsers Chrome![]() Firefox
Firefox ![]() or IE version (9,10,11)
or IE version (9,10,11) ![]()
The description of homepage is as follows
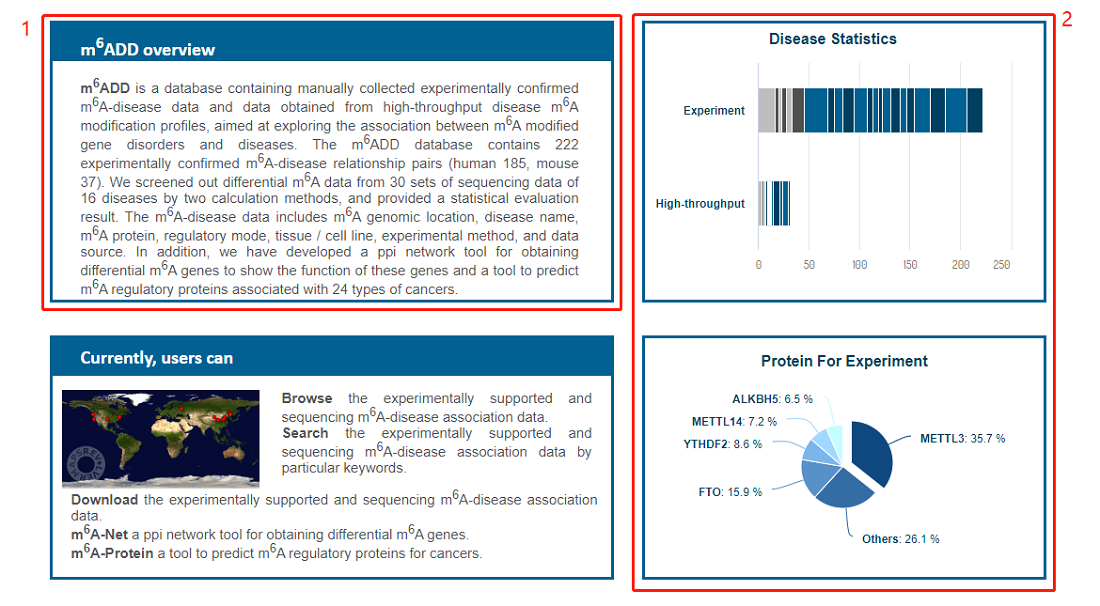
1. A simple introduction about the database
2. The statistics of the data in database, including disease and protein for experiment.
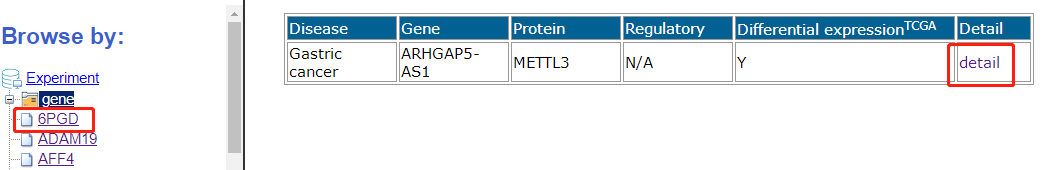
Users can browse entries based on gene, disease name and protein and click on the "Detail" column for more information.
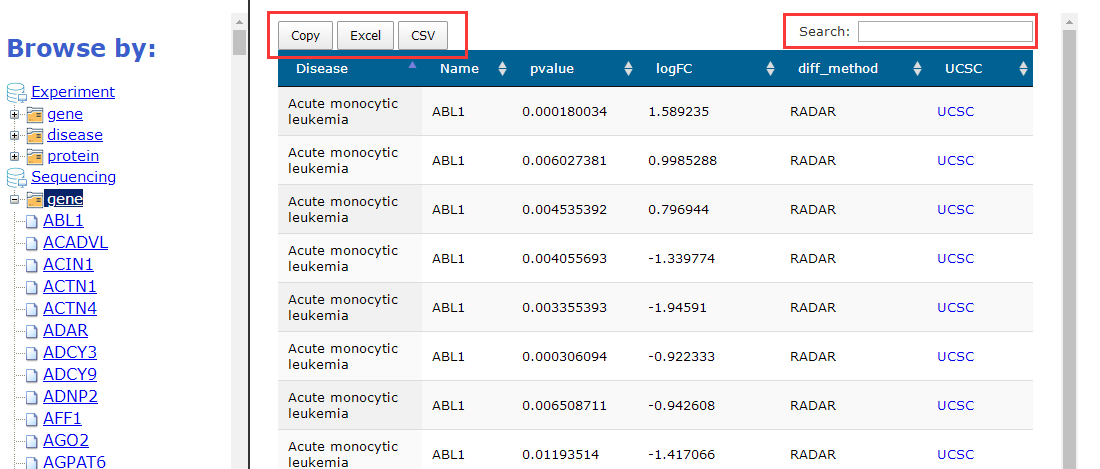
For sequencing data, we selected the hotspot differential m6A genes and put them in the browse interface, users can screen and export the results.
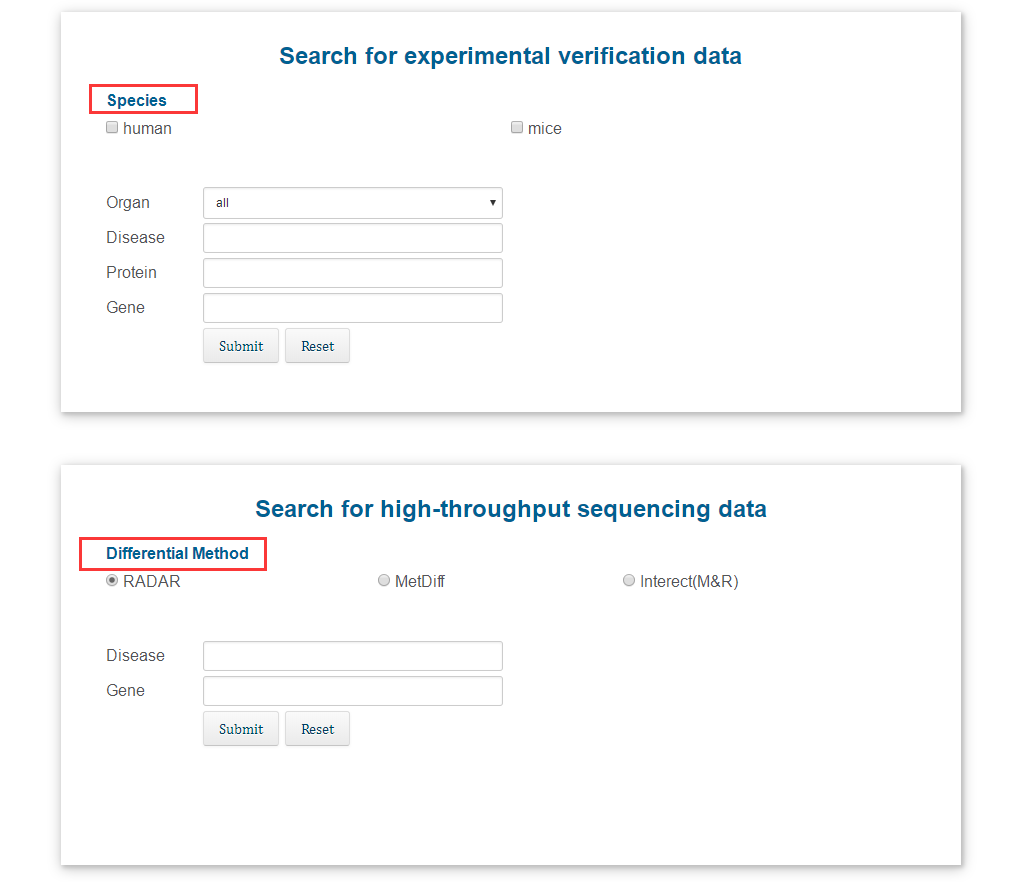
Users can search by entering keywords. For the "species" information, the user can select "human" or "mouse", or both. For the "difference method" information, the user can select a method to search. It should be noted that the result obtained by selecting "Intersect(M&R)" is based on sharing the same gene, while retaining the results of the two methods.
After entering a keyword to search experimental verification data , the database will provide a result interface. You can get the information of the data by clicking the "detail" button. For example, when you type keyword "MYC", you can get a page as follows
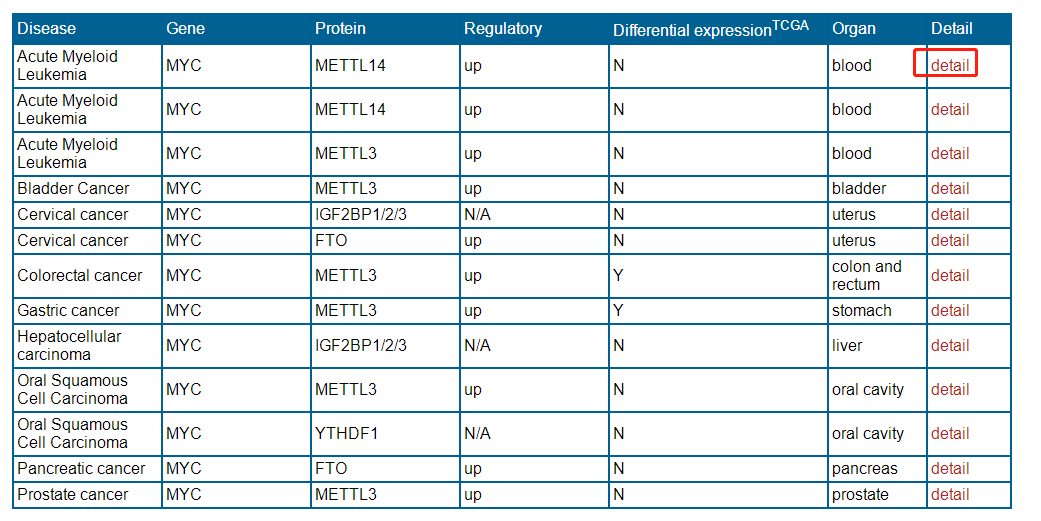
when you click on the "detail"
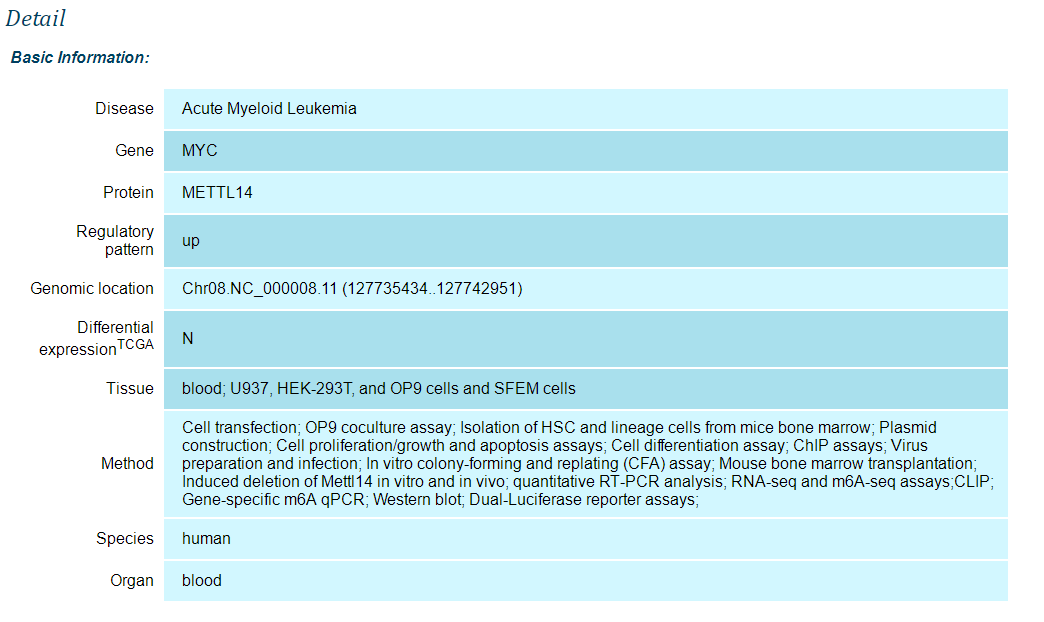

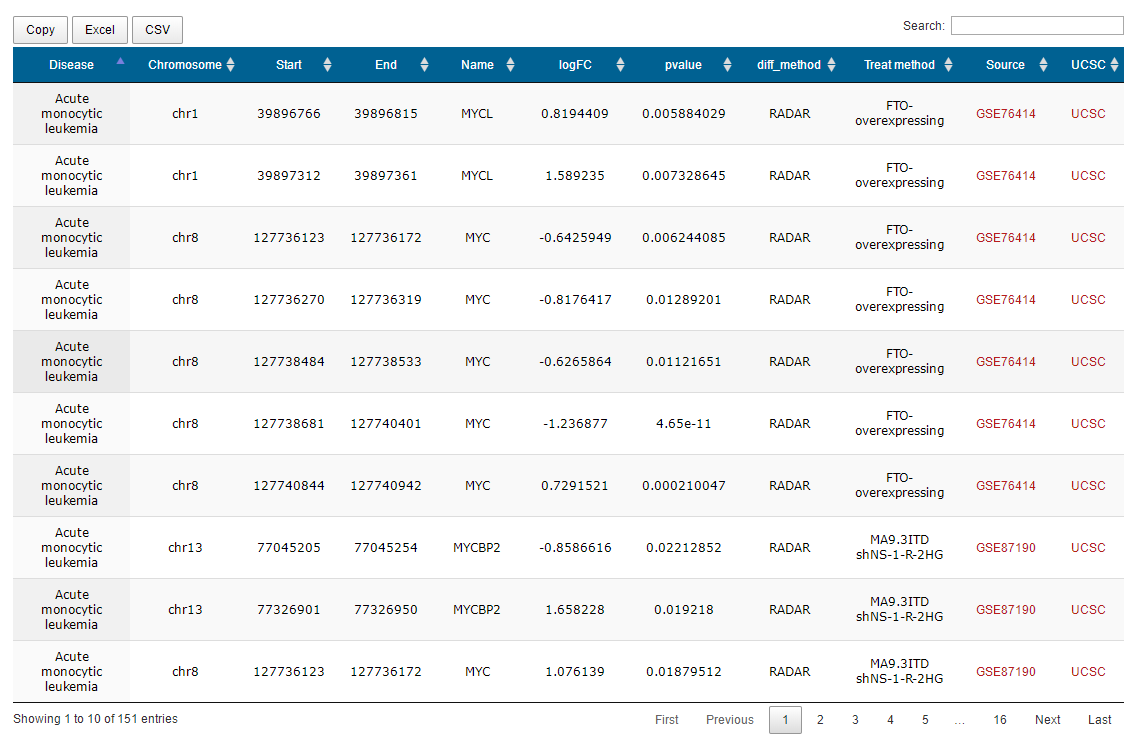
After entering a keyword to search high-throughput sequencing data , the database will provide a result interface. You can get the information of the data by clicking the "detail" button. For example, when you type keyword "MYC", you can get a page as follows
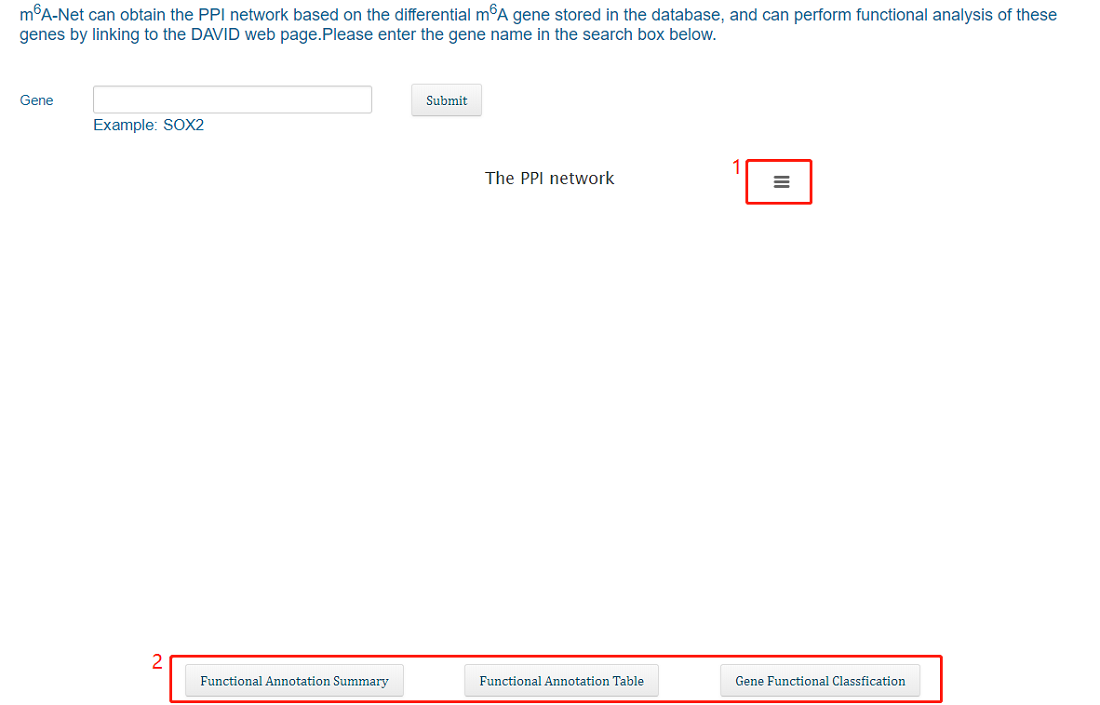
We get PPI data with a score greater than 900 from the string database. Users can get a PPI network interacting with this gene by entering a different m6A gene. Restricted by the page, the PPI network can display up to 30 nodes. Users can also click the button below to link to the "DAVID" website to perform functional annotation of the resulting gene set.
1. Users can export picture by clicking
2. DAVID website link.
By restructuring the PPI network, we predicted some genes that may perform similar functions to these three types of m6A regulatory factors in cancer. The user can obtain a list of genes with similar functions to the m6A regulatory factor in a specific cancer by selecting the type of cancer and the gene name of the m6A regulatory factor.

2020 © School of Life Science and Technology, Computational Biology Research Center | Harbin, 150001, Heilongjiang,P.R. China